Methods of Studying Viruses
For isolation and cultivation
1) Animals /Embryonated eggs
2) Plants
3) Tissue culture
For detection, identification and diagnosis
1) Tissue culture methods
2) Physical methods
a) X-ray crystallography
b) Electron-Microscopy
c) Ultra-centrifugation
3) Serological methods/Immunological methods
a) Haemagglutination (HA)
b) Haemagglutination Inhibitation (HI)
c) Complement Fixation
d) Immunostaining
e) Immunoprecipitation / Immunoblot
f) ELISA
4) Others and Molecular Biology
a) PAGE / SDS-PAGE
b) Western blot
c) Protein Sequencing
d) X-ray Crystallography
e) Agarose Gels
f) Restriction Analysis
g) Sequencing
h) Southern blot
i) Northen Blot
j) PCR / RT-PCR
METHODS FOR ISOLATION AND CULTIVATION
(1) Animals and Eggs
Using animals and embryonated eggs was one of the earliest methods to culture viruses. It is a rather inconvenient method as factors such as safety when handling animals and animal rights have to be considered. As a result in present day this method is mostly phased out and replaced by in vitro tissue culture methods.
However some viruses that has no known host in vitro and has to be cultured this way. Examples are the influenza virus, hepatitis C and polio.
In this method, animals and embryonated eggs are directly inoculated with the virus and the virus is allowed to replicate. The viruses cultured in these animals are then extracted.
(2) Plants
In plants viruses are also directly inoculated into them for cultivation and studying. For viruses such as the tobacco mosaic, the quantity of viruses in the plant can be estimated due to the ability of the tobacco mosaic virus to form plaques.
(3) Tissue culture
In the tissue culture method, the virus is grown in vitro in a culture of host cells, these viruses cultured can be used to infect other cultures of host cells to increase the virus titer.
In this method, a primary cell line is created and from it a continuous cell line is derived which will be infected with viruses to culture them. This is the method of choice for culturing viruses.
The general procedure is of creating continuous cell lines of animal cells are shown below:
1) First, remove the tissue to be cultured from a plant or animal.
2) Next, crush and grind the tissue into small pieces.
3) The crushed tissue is trypsinised
4) The trypsinised tissue is next placed into a culture medium and allowed to grow.
5) The primary cell line is created.
6) This primary cell line is treated with chemicals to transform it into cancerous cells that can keep dividing indefinitely.
7) This creates a continuous cell line which can be used to study viruses and culture them.
The general procedure of creating a plant tissue culture is shown below.
1) Cut-out plant tissue and place in a tissue culture container.
2) Tissue grows and produces small plants
3) Some are removed and placed in other culture containers to create a rapid multiplication of plant cultures.
4) When plants are large enough they are transplanted into soil and placed in a green house.
5) These plants are clones of the original plant and can be infected with plant viruses to culture them.
METHODS FOR DETECTION, IDENTIFICATION AND DIAGNOSIS
(1) Tissue Culture Methods
There are two methods for using tissue culture to detect viruses which are observing cytopathic effects and using the plaque assay.
Cytopathic Effects (CPE)
The presence of viruses can be detected by observing changes in the cell due to virus’s replication cycle.
Examples of the effects which can be observed in cells under the microscope are:
· Rounding of cells
· Development of a double nucleus.
· Cell lysis
· Swelling/Shrinking of Cells
· Detachment from surface
· Etc
CPEs vary depending on the type of virus infecting the cell.
Plaque Assay
Plaque assay is a method based on the principle that one virus will infect one cell which will produce a visible plaque on a cell monolayer in a plaque assay.
In a plaque assay the following takes place:
- Cell monolayers are infected with a low ratio of virus
- A layer of agarose on the cell monolayers keeps the cells stable and limits the spread of virus.
- When each infected cell produces virus and eventually lyses, only the cells immediately adjacent to it becomes infected.
- Each group of infected cells is referred to as a plaque.
- Uninfected cells surround the plaques.
- After several infection cycles, the infected cells in the center of the plaques begin to lyse and the surrounding infected cells remain surrounded by uninfected cells.
- Plaques can be seen by staining the monolayer.
- A areas of unstained regions are plaques and can be counted.
The plaque assay despite being a very simple method is very time-consuming and can only work for cells that can infect monolayers and cause cell lysis.
(2) Physical Methods
a) X-Ray Crystallography
In this method, X-rays are beamed at the crystal and electrons of the atoms of the sample being studied diffract the x-rays causing a diffraction pattern. Using mathematical formulae and computer programs 3-dimensional pictures and electron density maps can be created.
Since viruses are too small to be studied under a standard light-microscope, this method can be used to observe viruses.
b) Electron-Microscopy
Since viruses are too small to be observed in a light microscope, electron microscopy can be used to created detailed and accurate images of viruses. Electron-microscopy works by bombarding the specimen with a beam of electrons and the diffracted electrons can be used to form a image by using sensors connected to a computer that displays an image.
There are 2 types of electron microscopes:
· Transmission electron microscope (TEM)
· Scanning Electron microscope (STEM)
The image below shows the image produced by a TEM.
The image below shows the image produced by a STEM.
c) Ultra-centrifugation
Utra-centrifugation can be used to separate marcomolecules from each other when studying specimens such as cells or viruses. This enables researchers to obtain a purified sample of viruses.
Other function of ultra-centrifugation when using an analytical ultra-centrifuge are
- to monitor the number and molar mass of macromolecular complexes
- and to report on the shape and molar mass of the dissolved macromolecules, as well as their size-distribution.
A picture of a ultra-centrifuge is shown below.
(3) Serological methods/Immunological methods
a) Haemagglutination (HA)
Certain types of viruses such as the influenza virus contain spikes on their envelope which are called:
· Haemaglutanin
· Neuraminidase
The haemagglutanin spike binds to specifically to red blood cells bringing them together form clumps of red blood cells which will float to the surface of the liquid. Hence the no. of viruses can be enumerated by observing the amount of red blood cells clumped at the surface.
b) Haemagglutination Inhibitation (HI)
A clinical lab test used to detect the presence of a certain haemagglutinating virus or other haemagglutinin antigen based on whether the red blood cells in the sample lose the ability to clump together when the antibody to the virus or other antigen is added to it.
Haemagglutination Inhibition uses the same principle as ordinary Haemagglutination except that it uses antibody inhibition which binds to the virus neutralising it and hence preventing it from binding to red blood cells causing agglutination.
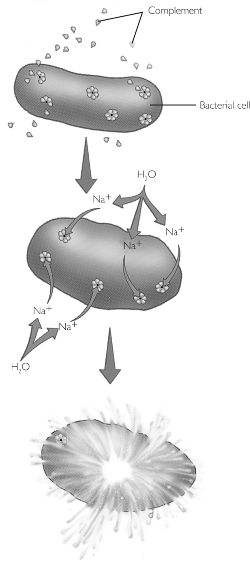
c) Complement Fixation
Complement fixation a a method where by an antibody binds to an antigen causing a complement cascade of molecules in the blood serum which interacts with the cell or pathogen causing it to lyse.
d) Immunostaining
Immunofluorescence
An antibody is tagged with fluorescent dye which attaches to a specific antigen of a sample. The sample is then observed under a fluorecent microscope which will produce exciting light to illuminate the fluorescent dye. This method can be used to study where and how the antibody and antigens react and bind together.
Immunogold Electron microscopy
Utilising the same principle in immunofluorescent staining except that flourescent dye is replaced with gold, antibody and antigen complexes can be observed in electron microscope where the gold dust can be seen.
e) Immunoprecipitation / Immunoblot
Immunoprecipitation
In this method a radioactively labelled antigen is reacted with an antibody creating a complex. This complex is will be run through SDS-PAGE and detected using an X-ray film. The diagram below furher explains this concept.
Immunoblot
A laboratory procedure, such as Western blot analysis, in which proteins that have been separated by electrophoresis are transferred onto nitrocellulose sheets and are identified by their reaction with labeled antibodies.
In this western blot analysis, the whole protein to be studied is run through SDS-PAGE and blotted onto nitrocellulose paper. Antibodies labelled with an indicator added to the paper, bind to the proteins on it. The paper is then treated with streptavidin which reacts with the indicator producing a colour reaction.
This diagram shows how the western blot is done. But in the case above radioactively labelled antibodies are used.
f) ELISA
ELISA also known as enzyme-linked immunosorbent assay is a biocemical technique used in immunology in the detection or antigens or antibodies that may be present in the sample.
One way the ELISA can work is by allowing an antibody to bind to the antigen on the well’s surface if the correct antibody-antigen pair is present. After which an enzyme conjugate added binds to the antibody-antigen complex, activatin the enzyme which converts an added substrate into a coloured compound. Any component missing will result in a negative test.
The diagram below explains this concept.
(4) Others and Molecular Biology
a) PAGE / SDS-PAGE
PAGE or Polyacrylamide Gel Electrophoresis is a method whereby proteins are separated by using an electric field to separate them according to their sizes in a polyacrylamide gel medium. The electric field produced pull the proteins through the ployacrylamide gel which is composed of a laberynth of tunnels within a mesh of fibres.
The SDS in SDS-PAGE is the step done before PAGE whereby the proteins are denatured so that it is reduced to its primary structure and that all proteins’ 3D shapes do not affect their rate of movement in the polyacrylamide gel in PAGE.
The SDS is actually an acronym for sodium dodecyl sulfate which is the detergent used to denature the proteins. The detergent dissolves hydrophoic molecules and has a negative charge attached to it. So if it is used on a protein, it dissolves it, denatures it and gives it negative charges. Hence in PAGE, proteins will migrate to the positive pole.
The diagram below shows what happens when proteins are degraded by SDS.
b) Western blot
This was discussed in the earlier section on immunoblot.
c) Protein Sequencing
In protein sequencing, the amino acid sequence on the poly peptide’s chain is determined by various methods.
To determine the composition of the protein the following can be done.
- First, the proteins sample is hydrolysed into its constituent amino acid by heating in hydrochloric acid.
- Next, the proteins are separated by ion-exchange or hydrophobic interaction chromatography.
- The separated amino acids will next undergo quantitative analysis producing colour changes.
- These colour changes in each separated amino acid can be put into a photospectrometer to obtain its corresponding absorbances.
- These absorbances are directly related to the amount ofamino acid present hence the composition of the amino acid can be determined.
Another method in protein sequence is mass spectrometry where by the specific amino acid sequences can be determined. Mass spectrometry work by ionising the sample to created charged ions where their mass to charge ratio is determined. This ratio is calculated from the motion of the ions as they pass through electromagnetic fields.
The pictures below show a mass spectrometer and the data produced by it.
d) X-ray Crystallography
This was discussed earlier under the physical methods section.
e) Restriction Analysis
In the restriction analysis method the genetic material for example DNA is cut into segments using restriction enzymes and run through PAGE or gel electrophoresis separating the segments which can be analysed.
f) DNA Sequencing
In DNA sequencing, a mixture of bases, enzymes and cofactors are reacted with a DNA strand to create a complementary strand which can be run through gel electrophoresis. Through gel electroporesis the DNA bases can be sorted according to sequence and base pair type. By comparing the results with complementary bases, the sequence of the sample DNA strand can be recontructed.
g) Southern blot
Southern blot is a technique developed for transferring DNA fragments from gel electrophoresis onto the nitrocellulose paper. This is essential so that the DNA fragments can be detected using techniques such as labelling fragments with radioactive material.
The picture below shows how sourthern blotting is done.
h) Northen Blot
The northern blot analysis is based on the same principle as with the southern blot except that northern blot is used for RNA instead.
i) PCR / RT-PCR
PCR
PCR also known as Polymerase Chain Reaction is used to create a large amount of copies of DNA. This is useful especially when the DNA sample being studied is low in quantity.
PCR works by repeating a cycle of 3 steps over and over again till enough duplicate DNA is produced. The steps are:
- First, the double stranded DNA is heated to separate the DNA into single strands.
- Next, primers and binded to DNA strand upon cooling to designate the area of the DNA strand to be duplicated.
- Lastly, temperature is again increased allowing the polymerase enzyme to attach the bases to the DNA strand creating 2 copies of the same DNA.
RT-PCR
RT-PCR is used for RNA and works almost the same as standard PCR only with a an additonal step.
RT-PCR works by first converting RNA into DNA using reverse transcriptase to produce a complementary strand. Next the standard 3 step PCR cycle is used to create duplicates.
The video below shows how PCR works.
http://www.youtube.com/watch?v=j9Gu7iwBi4I
References:
1) http://generalhorticulture.tamu.edu/YouthAdventureProgram/TisueCulture/P81F1.GIF
2) http://www.scielosp.org/img/revistas/rsp/v34n4/2532f2.jpg
3) http://pathmicro.med.sc.edu/mhunt/plaque.jpg
4) http://porpax.bio.miami.edu/~cmallery/255/255tech/mcb3.38.xray.jpg
5) http://www3.imperial.ac.uk/pls/portallive/docs/1/872005.JPG
6) http://www.sci.sdsu.edu/emfacility/Images/temimage.JPG
7) http://medschool.umaryland.edu/infeMSD/wtlasema.JPG
8) http://www.id.yamagata-u.ac.jp/CLRE/img/biochem_gif/u-cent-mini-top.gif
9) http://www.umanitoba.ca/science/microbiology/staff/cameron/graphics/401lab3directhemagglutination%20group106.jpg
10) http://img.tfd.com/mosbycam/thumbs/500069-fx32.jpg
11) http://www.abcam.com/ps/datasheet/images/Ab31531_2.gif
12) http://www.piercenet.com/media/fig1profound.gif
13) http://microvet.arizona.edu/Courses/MIC419/ToolBox/ELISA2.gif
14) http://www.bio.davidson.edu/COURSES/GENOMICS/method/SDSPAGE/SDSwprotein.GIF
15) http://upload.wikimedia.org/wikipedia/commons/5/53/Pnnl_ftms.jpg
16) http://upload.wikimedia.org/wikipedia/commons/7/7b/ObwiedniaPeptydu.gif
17) http://sg.wrs.yahoo.com/_ylt=A0S0zu5ruYZJ7WsBifQu4gt./SIG=128t79avc/EXP=1233652459/**http%3A/www.science2discover.com/images/southern2.gif
Information:
1) http://pathmicro.med.sc.edu/mhunt/replicat.htm
2) http://www.stolaf.edu/people/hansonr/mo/x-ray.html
3) http://www.bio.davidson.edu/COURSES/GENOMICS/method/SDSPAGE/SDSPAGE.html
6:05 PM